Objectives: learn how to define outputs and create tables from the outputs of the model.
Projects: tgi_project, tgiWithTable_project
About the OUTPUT block
- tgi_project (data = ‘tgi_data.txt’ , model=’tgi_model.txt’)
We use the Tumor Growth Inhibition (TGI) model proposed by Ribba et al. in this example (Ribba, B., Kaloshi, G., Peyre, M., Ricard, D., Calvez, V., Tod, M., . & Ducray, F., A tumor growth inhibition model for low-grade glioma treated with chemotherapy or radiotherapy. Clinical Cancer Research, 18(18), 5071-5080, 2012.)
DESCRIPTION: Tumor Growth Inhibition (TGI) model proposed by Ribba et al
A tumor growth inhibition model for low-grade glioma treated with chemotherapy or radiotherapy.
Clinical Cancer Research, 18(18), 5071-5080, 2012.
Variables
- PT: proliferative equiescent tissue
- QT: nonproliferative equiescent tissue
- QP: damaged quiescent cells
- C: concentration of a virtual drug encompassing the 3 chemotherapeutic components of the PCV regimen
Parameters
- K : maximal tumor size (should be fixed a priori)
- KDE : the rate constant for the decay of the PCV concentration in plasma
- kPQ : the rate constant for transition from proliferation to quiescence
- kQpP : the rate constant for transfer from damaged quiescent tissue to proliferative tissue
- lambdaP: the rate constant of growth for the proliferative tissue
- gamma : the rate of damages in proliferative and quiescent tissue
- deltaQP: the rate constant for elimination of the damaged quiescent tissue
- PT0 : initial proliferative equiescent tissue
- QT0 : initial nonproliferative equiescent tissue
[LONGITUDINAL]
input = {K, KDE, kPQ, kQpP, lambdaP, gamma, deltaQP, PT0, QT0}
PK:
depot(target=C)
EQUATION:
; Initial conditions
t0 = 0
C_0 = 0
PT_0 = PT0
QT_0 = QT0
QP_0 = 0
; Dynamical model
PSTAR = PT + QT + QP
ddt_C = -KDE*C
ddt_PT = lambdaP*PT*(1-PSTAR/K) + kQpP*QP - kPQ*PT - gamma*KDE*PT*C
ddt_QT = kPQ*PT - gamma*KDE*QT*C
ddt_QP = gamma*KDE*QT*C - kQpP*QP - deltaQP*QP
OUTPUT:
output = PSTAR
PSTAR is the tumor size predicted by the model. It is therefore used as a prediction for the observations in the project.
At the end of the scenario or of SAEM, individual predictions of the tumor size PSTAR are computed using the individual parameters available. Thus, individual predictions of the tumor size PSTAR are computed using both the conditional modes (indPred_mode), the conditional mean (indPred_mean), and the conditional means estimated during the last iterations of SAEM (indPred_SAEM) and saved in the table predictions.txt.
Notice that the population prediction is also proposed.

Remark: the same model file tgi_model.txt can be used with different tools, including Mlxplore or Simulx.
Add additional outputs in tables
- tgiWithTable_project (data = ‘tgi_data.txt’ , model=’tgiWithTable_model.txt’)
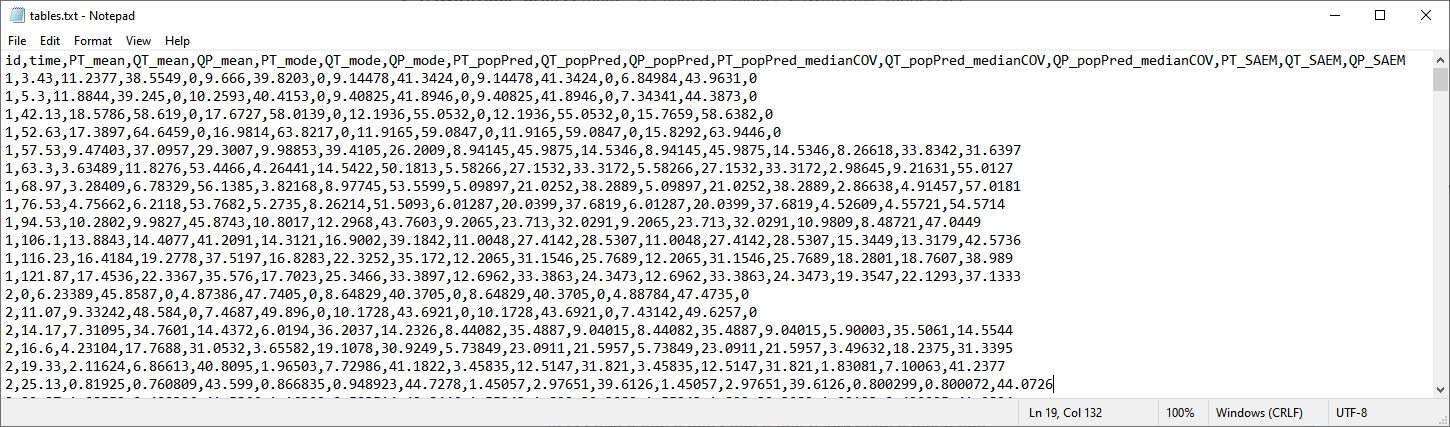
We can save in the tables additional variables defined in the model, such as PT, Q and QP for instance, by adding a block OUTPUT: in the model file:
OUTPUT:
output = PSTAR
table = {PT, QT, QP}
An additional file tables.txt now includes the predicted values of these variables for each individual (columns PT_mean, QT_mean, QP_mean, PT_mode, QT_mode, QP_mode, PT_popPred, QT_popPred, QP_popPred, PT_popPred_medianCOV, QT_popPred_medianCOV, QP_popPred_medianCOV, PT_SAEM, QT_SAEM, and QP_SAEM.
Notice that only continuous variable are possible for variable in table.
Good to know: it is not allowed to do calculations directly in the output or table statement. The following example is not possible:
; not allowed:
OUTPUT:
output = {Cser+Ccsf}
It has to be replaced by:
EQUATION:
Ctot = Cser+Ccsf
OUTPUT:
output = {Ctot}