Download data set only | Monolix project files | Simulx project files and scripts
[project files coming soon]
⚠️ Under construction ⚠️
This case study presents modeling of the tobramycin pharmacokinetics and determination of a priori dosing regimens in patients with various degrees of renal function impairment. It takes advantage of the integrated use of Monolix for data visualization and parameter estimation and Simulx for simulations and best dosing regimen determination. The case study is presented in 4 sequential parts which we recommend reading in order:- Part 1: Introduction
- Part 2: Exploratory Data Analysis with Monolix
- Part 3: Model development with Monolix
- Part 4: Simulations for individualized dosing with Simulx and Monolix
Part 2: Exploratory data analysis with Monolix
Data set overview
Tobramycin bolus doses ranging from 20 to 140mg were administrated every 8 hours in 97 patients (45 females, 52 male) for 1 to 21 days (for most patients, for ~6 days). Age, weight (kg), sex, height and creatinine clearance (mL/min) are available as covariates. Because height information was missing for around 30% of the patients, this covariate is ignored. The tobramycin concentration (mg/L) was measured 1 to 9 times per patients (322 measures in total), generally between 2 and 6h post-dose. Below is an extract of the data set file: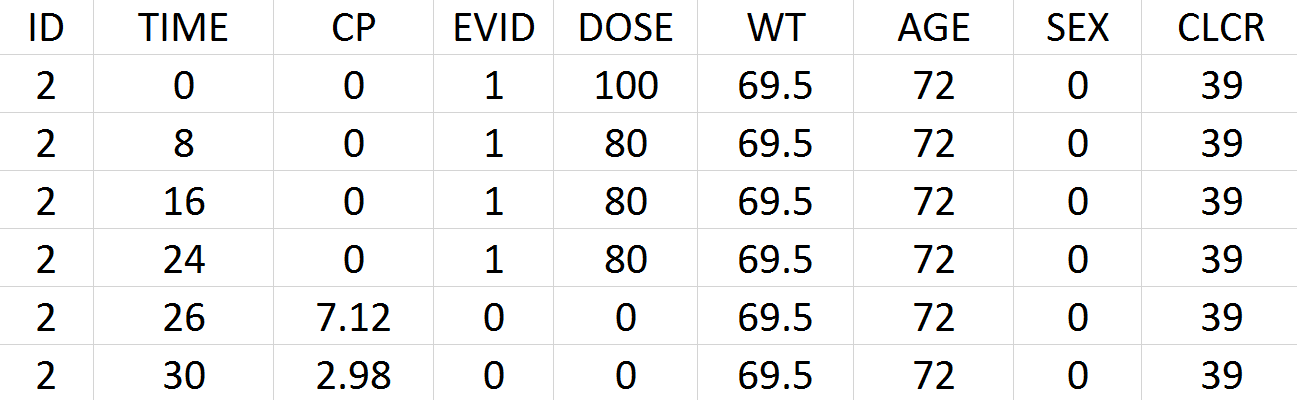
- ID: identifier of the patients
- TIME: time of dose or measurement (hours)
- CP: Tobramycin plasma concentration (mg/L)
- EVID: event identifier, 0=measurement, 1=dose
- DOSE: amount of the dose (mg)
- WT: weight (kg)
- AGE: age (years)
- SEX: 1=male, 0=female
- CLCR: creatinine clearance (mL/min)
Data set visualization with Monolix
We first start exploring the data set in Monolix. After having opened Monolix, we create a new project and load the data set. Based on the header, most columns are automatically recognized. The column-type must be set manually for the observed concentration (CP column set to column-type OBSERVATION), the dose (DOSE column set to column-type AMOUNT), and the creatinine clearance (CLCR column set to column-type CONTINUOUS COVARIATE for continuous covariate):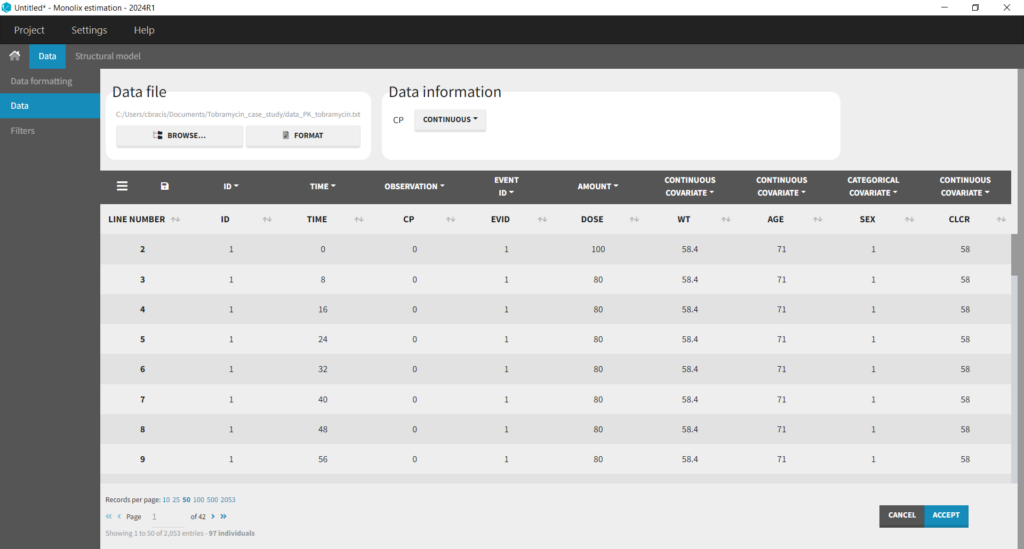
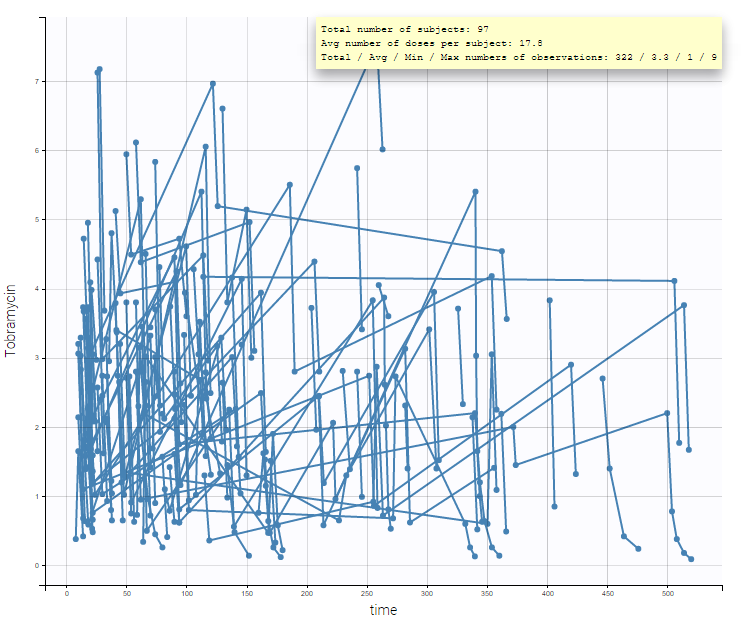
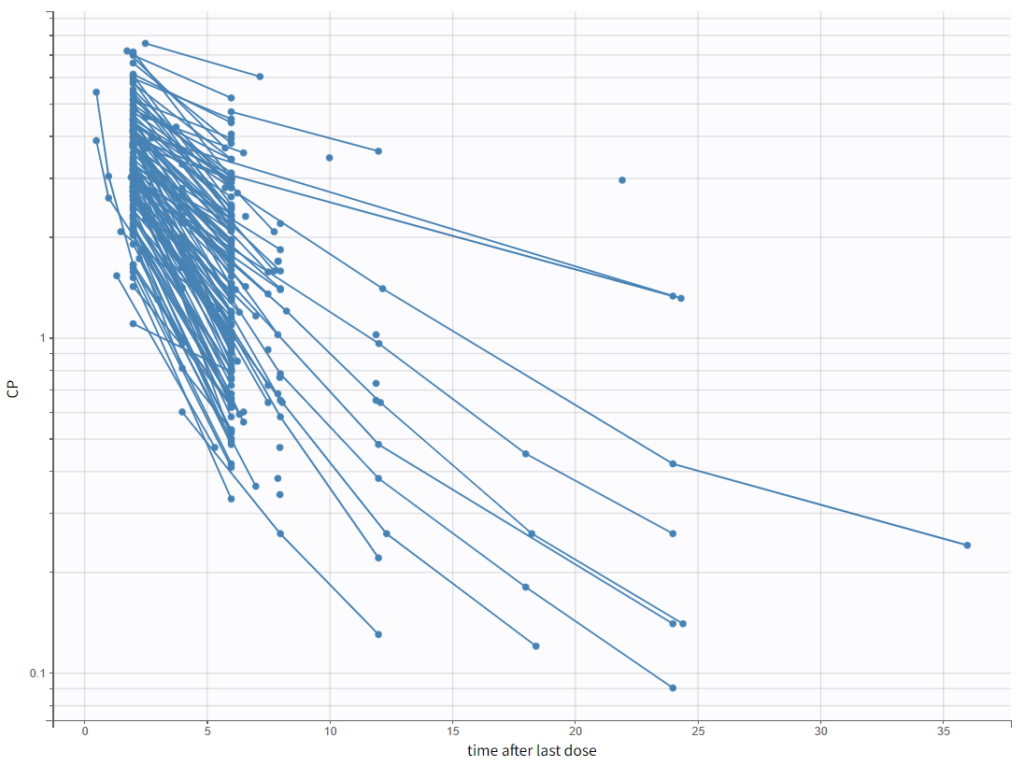
- A 1 compartment model with linear elimination is a good first approximation, and we can also explore a 2 compartment model.
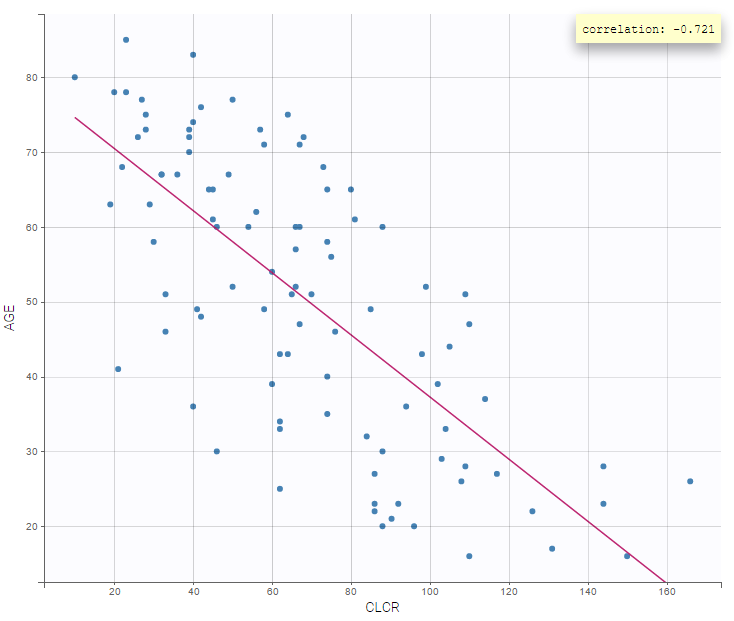
- Older patients have a lower creatinine clearance.